Google ASL 1. Process Data with W&B 🐝
Today, I’m going to explain the dataset and how to process it for a Kaggle competition on ASL(American Sign Language), Google — Isolated…
Google ISLR Transformer with W&B (Part 1)
In this Medium article, I’ll guide you through the process of preparing a landmark dataset for a Kaggle competition on Google — Isolated Sign Language Recognition. The competition’s task is to classify isolated signs using. ASL is a complete and natural language that uses hand gestures, facial expressions, and body language to communicate. Machine learning can help us develop models that accurately and efficiently recognize ASL signs. Throughout this article, I’ll provide you with a step-by-step guide on how to process the dataset and make it ready for model training. So, let’s dive into the world of ASL and get started!
I used and modified Mark Wijkhuizen’s code, and monitored the training using W&B. You can check the full code and my W&B dashboard.
I have downloaded the dataset below and uploaded it to Google Drive.
- https://www.kaggle.com/competitions/asl-signs/data
- https://www.kaggle.com/datasets/markwijkhuizen/gislr-dataset-public
gislr-dataset-public has 6 types of datasets to be processed:
- X: A 4D numpy array of shape [N_SAMPLES, INPUT_SIZE, N_COLS, N_DIMS] containing the preprocessed data for all samples.
- y: A 1D numpy array of shape [N_SAMPLES] containing the corresponding target labels for all samples.
- NON_EMPTY_FRAME_IDXS: A 2D numpy array of shape [N_SAMPLES, INPUT_SIZE] containing the indices of non-empty frames for all samples.
- X_train/X_val: 4D numpy arrays of shape [N_TRAIN_SAMPLES, INPUT_SIZE, N_COLS, N_DIMS] containing the preprocessed data for the training/validation sets.
- y_train/y_val: 1D numpy arrays of shape [N_TRAIN_SAMPLES] containing the corresponding target labels for the training/validation sets.
- NON_EMPTY_FRAME_IDXS_TRAIN/NON_EMPTY_FRAME_IDXS_VAL: 2D numpy arrays of shape [N_TRAIN_SAMPLES, INPUT_SIZE] containing the indices of non-empty frames for the training/validation sets.
Imports
import numpy as np
import pandas as pd
import tensorflow as tf
import tensorflow_addons as tfa
import tflite_runtime.interpreter as tflite
import matplotlib.pyplot as plt
import matplotlib as mpl
import seaborn as sn
import wandb
from tqdm.notebook import tqdm
from sklearn.model_selection import train_test_split, GroupShuffleSplit
import glob
import sys
import os
import math
import gc
import sys
import sklearn
import scipy
from types import SimpleNamespace
# Vesion Check
print(f'Tensorflow V{tf.__version__}')
print(f'Keras V{tf.keras.__version__}')
print(f'Python V{sys.version}')
'''
Tensorflow V2.12.0
Keras V2.12.0
Python V3.9.16 (main, Dec 7 2022, 01:11:51)
'''- numpy: A scientific computing library that provides multidimensional array and matrix data structures as well as various mathematical functions to operate on them. pandas: A data analysis and manipulation library that provides data structures and operations for numerical tables and time series.
- tensorflow: An open-source machine learning platform that provides a comprehensive and flexible framework for developing and deploying various types of neural networks. tensorflow_addons: A contributed library that follows some mature API patterns, but implements new features that are not available in core TensorFlow.
- matplotlib: A plotting library that can generate publication-quality graphics in a variety of formats and interactive environments.
- seaborn: A data visualization library based on matplotlib that provides a high-level interface for creating aesthetically pleasing and informative statistical graphics.
- tqdm: A fast and extensible Python and CLI progress bar.
- sklearn: A machine learning library that provides various classification, regression, clustering, dimensionality reduction, model selection, and preprocessing algorithms. glob: A module that provides a function for generating a list of files that match a given pattern.
- sys: A module that provides an interface for accessing some variables and functions used or maintained by the interpreter.
- os: A module that provides a portable way of using operating system-dependent functionality.
- math: A module that provides an interface to the C standard math functions.
- gc: A module that provides an interface for accessing the optional garbage collector.
- scipy: A scientific computing library that provides many user-friendly and efficient numerical routines such as numerical integration, interpolation, optimization, linear algebra, and statistics.
Global Config & Utils
Since the data has already been processed, PREPROCESS_DATA is set as False.
cfg = SimpleNamespace()
# If True, processing data from scratch
# If False, loads preprocessed data
cfg.PREPROCESS_DATA = False
cfg.TRAIN_MODEL = True
# True: use 10% of participants as validation set
# False: use all data for training
cfg.USE_VAL = False
cfg.N_ROWS = 543
cfg.N_DIMS = 3
cfg.DIM_NAMES = ['x', 'y', 'z']
cfg.SEED = 42
cfg.NUM_CLASSES = 250
cfg.IS_INTERACTIVE = True
cfg.VERBOSE = 2
cfg.INPUT_SIZE = 64
cfg.BATCH_ALL_SIGNS_N = 4
cfg.BATCH_SIZE = 256
cfg.N_EPOCHS = 100
cfg.LR_MAX = 1e-3
cfg.N_WARMUP_EPOCHS = 0
# Dropout Ratio
cfg.WD_RATIO = 0.05
#cfg.MASK_VAL = 4237# Make a project on W&B 🐝
if cfg.TRAIN_MODEL:
run = wandb.init(project="kaggle-asl-signs", config=cfg, tags=['transformer', 'final-model'])# MatplotLib Global Settings
mpl.rcParams.update(mpl.rcParamsDefault)
mpl.rcParams['xtick.labelsize'] = 16
mpl.rcParams['ytick.labelsize'] = 16
mpl.rcParams['axes.labelsize'] = 18
mpl.rcParams['axes.titlesize'] = 24# Prints Shape and Dtype For List Of Variables
def print_shape_dtype(l, names):
for e, n in zip(l, names):
print(f'{n} shape: {e.shape}, dtype: {e.dtype}')Read Training Data
# Read Training Data
BASE = '/content/drive/MyDrive/23_Google/data'
# if PREPROCESS_DATA is fale, N_SAMPLES is 5,000
if not cfg.PREPROCESS_DATA:
train = pd.read_csv(f'{BASE}/train.csv').sample(int(5e3), random_state=cfg.SEED)
else:
train = pd.read_csv(f'{BASE}/train.csv')
N_SAMPLES = len(train)
print(f'N_SAMPLES: {N_SAMPLES}')
# Get complete file path to file
def get_file_path(path):
return f'{BASE}/{path}'
train['file_path'] = train['path'].apply(get_file_path)Ordinally Encode Sign
The sign column of the train is encoded ordinally, assigning a number to each sign name.
# Add ordinally Encoded Sign (assign number to each sign name)
train['sign_ord'] = train['sign'].astype('category').cat.codes
# Dictionaries to translate sign <-> ordinal encoded sign
SIGN2ORD = train[['sign', 'sign_ord']].set_index('sign').squeeze().to_dict()
ORD2SIGN = train[['sign_ord', 'sign']].set_index('sign_ord').squeeze().to_dict()
display(train.head(30))
display(train.info())Landmark Indices
There are 3 types of landmarks. There are Keypoints used in MediaPipe Facemesh so we use predetermined LIP Keypoints. The landmark indices for the corresponding body parts are concatenated and are used to extract the relevant landmarks from the data. N_COLS is the total number of landmarks.
# Define three data types: left hand, pose, and right hand
SE_TYPES = ['left_hand', 'pose', 'right_hand']
# Define starting index in original data
START_IDX = 468
# Define lip landmark indexes in original data, a total of 40
LIPS_IDXS0 = np.array([
61, 185, 40, 39, 37, 0, 267, 269, 270, 409,
291, 146, 91, 181, 84, 17, 314, 405, 321, 375,
78, 191, 80, 81, 82, 13, 312, 311, 310, 415,
95, 88, 178, 87, 14, 317, 402, 318, 324, 308,
])
# Landmark indices in original data
# Define left hand landmark indexes in original data, a total of 21
LEFT_HAND_IDXS0 = np.arange(468,489)
# Define right hand landmark indexes in original data, a total of 21
RIGHT_HAND_IDXS0 = np.arange(522,543)
# Define left pose landmark indexes in original data, a total of 5
LEFT_POSE_IDXS0 = np.array([502, 504, 506, 508, 510])
# Define right pose landmark indexes in original data, a total of 5
RIGHT_POSE_IDXS0 = np.array([503, 505, 507, 509, 511])
# Define landmark indexes for left-hand-dominant data, including lips, left hand, and left pose, a total of 66
LANDMARK_IDXS_LEFT_DOMINANT0 = np.concatenate((LIPS_IDXS0, LEFT_HAND_IDXS0, LEFT_POSE_IDXS0))
LANDMARK_IDXS_RIGHT_DOMINANT0 = np.concatenate((LIPS_IDXS0, RIGHT_HAND_IDXS0, RIGHT_POSE_IDXS0))
HAND_IDXS0 = np.concatenate((LEFT_HAND_IDXS0, RIGHT_HAND_IDXS0), axis=0)
# Define number of columns in processed data, which is 66
N_COLS = LANDMARK_IDXS_LEFT_DOMINANT0.sizenp.isin() returns a boolean array of the same shape as the first input array, indicating whether each element of the first array is also present in the second input array. np.argwhere() returns an array of indices where a specified condition is true.
np.isin() is used to create a boolean array indicating whether each landmark index in LANDMARK_IDXS_LEFT_DOMINANT0 is also present in one of the specified body parts or pose arrays. np.argwhere() is then used to return an array of the indices of the True elements in this boolean array. The resulting array of indices corresponds to the landmarks that belong to the specified body part or pose.
# Landmark indices in processed data
LIPS_IDXS = np.argwhere(np.isin(LANDMARK_IDXS_LEFT_DOMINANT0, LIPS_IDXS0)).squeeze()
LEFT_HAND_IDXS = np.argwhere(np.isin(LANDMARK_IDXS_LEFT_DOMINANT0, LEFT_HAND_IDXS0)).squeeze()
RIGHT_HAND_IDXS = np.argwhere(np.isin(LANDMARK_IDXS_LEFT_DOMINANT0, RIGHT_HAND_IDXS0)).squeeze()
HAND_IDXS = np.argwhere(np.isin(LANDMARK_IDXS_LEFT_DOMINANT0, HAND_IDXS0)).squeeze()
POSE_IDXS = np.argwhere(np.isin(LANDMARK_IDXS_LEFT_DOMINANT0, LEFT_POSE_IDXS0)).squeeze()
print(f'# HAND_IDXS: {len(HAND_IDXS)}, N_COLS: {N_COLS}')
# HAND_IDXS: 21, N_COLS: 66
LIPS_START = 0
LEFT_HAND_START = LIPS_IDXS.size
RIGHT_HAND_START = LEFT_HAND_START + LEFT_HAND_IDXS.size
POSE_START = RIGHT_HAND_START + RIGHT_HAND_IDXS.size
print(f'LIPS_START: {LIPS_START}, LEFT_HAND_START: {LEFT_HAND_START}, RIGHT_HAND_START: {RIGHT_HAND_START}, POSE_START: {POSE_START}')
# LIPS_START: 0, LEFT_HAND_START: 40, RIGHT_HAND_START: 61, POSE_START: 61Process Data Tensorflow
PreprocessLayer is TensorFlow layer designed to process data in TFLite. The purpose of this layer is to process the input data in a specific way that is required by the model. The processing steps involve filtering frames that contain data related to the dominant hand, normalizing the coordinates of the dominant hand, padding the data to a specific size, and filling NaN values with zeros.
The layer contains a call method that takes in a data0 tensor with shape [None, cfg.N_ROWS, cfg.N_DIMS] and processes it to return a tensor data with shape [cfg.INPUT_SIZE, cfg.N_ROWS, cfg.N_DIMS] and a tensor non_empty_frames_idxs with shape [cfg.INPUT_SIZE].
“Dominant” means the most frequently occurring element in the list.
"""
Tensorflow layer to process data in TFLite
Data needs to be processed in the model itself, so we can not use Python
"""
class PreprocessLayer(tf.keras.layers.Layer):
def __init__(self):
super(PreprocessLayer, self).__init__()
normalisation_correction = tf.constant([
# Add 0.50 to left hand (original right hand) and substract 0.50 of right hand (original left hand)
[0] * len(LIPS_IDXS) + [0.50] * len(LEFT_HAND_IDXS) + [0.50] * len(POSE_IDXS),
# Y coordinates stay intact
[0] * len(LANDMARK_IDXS_LEFT_DOMINANT0),
# Z coordinates stay intact
[0] * len(LANDMARK_IDXS_LEFT_DOMINANT0),
],
dtype=tf.float32,
)
self.normalisation_correction = tf.transpose(normalisation_correction, [1,0])
def pad_edge(self, t, repeats, side):
if side == 'LEFT':
return tf.concat((tf.repeat(t[:1], repeats=repeats, axis=0), t), axis=0)
elif side == 'RIGHT':
return tf.concat((t, tf.repeat(t[-1:], repeats=repeats, axis=0)), axis=0)
@tf.function(
input_signature=(tf.TensorSpec(shape=[None,cfg.N_ROWS,cfg.N_DIMS], dtype=tf.float32),),
)
def call(self, data0):
# Number of Frames in Video
N_FRAMES0 = tf.shape(data0)[0]
# Find dominant hand by comparing summed absolute coordinates
left_hand_sum = tf.math.reduce_sum(tf.where(tf.math.is_nan(tf.gather(data0, LEFT_HAND_IDXS0, axis=1)), 0, 1))
right_hand_sum = tf.math.reduce_sum(tf.where(tf.math.is_nan(tf.gather(data0, RIGHT_HAND_IDXS0, axis=1)), 0, 1))
left_dominant = left_hand_sum >= right_hand_sum
# Count non NaN Hand values in each frame for the dominant hand
if left_dominant:
frames_hands_non_nan_sum = tf.math.reduce_sum(
tf.where(tf.math.is_nan(tf.gather(data0, LEFT_HAND_IDXS0, axis=1)), 0, 1),
axis=[1, 2],
)
else:
frames_hands_non_nan_sum = tf.math.reduce_sum(
tf.where(tf.math.is_nan(tf.gather(data0, RIGHT_HAND_IDXS0, axis=1)), 0, 1),
axis=[1, 2],
)
# Find frames indices with coordinates of dominant hand
non_empty_frames_idxs = tf.where(frames_hands_non_nan_sum > 0)
non_empty_frames_idxs = tf.squeeze(non_empty_frames_idxs, axis=1)
# Filter frames
data = tf.gather(data0, non_empty_frames_idxs, axis=0)
# Cast Indices in float32 to be compatible with Tensorflow Lite
non_empty_frames_idxs = tf.cast(non_empty_frames_idxs, tf.float32)
# Normalize to start with 0
non_empty_frames_idxs -= tf.reduce_min(non_empty_frames_idxs)
# Number of Frames in Filtered Video
N_FRAMES = tf.shape(data)[0]
# Gather Relevant Landmark Columns
if left_dominant:
data = tf.gather(data, LANDMARK_IDXS_LEFT_DOMINANT0, axis=1)
else:
data = tf.gather(data, LANDMARK_IDXS_RIGHT_DOMINANT0, axis=1)
data = (
self.normalisation_correction + (
(data - self.normalisation_correction) * tf.where(self.normalisation_correction != 0, -1.0, 1.0))
)
# Video fits in INPUT_SIZE
if N_FRAMES < cfg.INPUT_SIZE:
# Pad With -1 to indicate padding
non_empty_frames_idxs = tf.pad(non_empty_frames_idxs, [[0, cfg.INPUT_SIZE-N_FRAMES]], constant_values=-1)
# Pad Data With Zeros
data = tf.pad(data, [[0, cfg.INPUT_SIZE-N_FRAMES], [0,0], [0,0]], constant_values=0)
# Fill NaN Values With 0
data = tf.where(tf.math.is_nan(data), 0.0, data)
return data, non_empty_frames_idxs
# Video needs to be downsampled to INPUT_SIZE
else:
# Repeat
if N_FRAMES < cfg.INPUT_SIZE**2:
repeats = tf.math.floordiv(cfg.INPUT_SIZE * cfg.INPUT_SIZE, N_FRAMES0)
data = tf.repeat(data, repeats=repeats, axis=0)
non_empty_frames_idxs = tf.repeat(non_empty_frames_idxs, repeats=repeats, axis=0)
# Pad To Multiple Of Input Size
pool_size = tf.math.floordiv(len(data), cfg.INPUT_SIZE)
if tf.math.mod(len(data), cfg.INPUT_SIZE) > 0:
pool_size += 1
if pool_size == 1:
pad_size = (pool_size * cfg.INPUT_SIZE) - len(data)
else:
pad_size = (pool_size * cfg.INPUT_SIZE) % len(data)
# Pad Start/End with Start/End value
pad_left = tf.math.floordiv(pad_size, 2) + tf.math.floordiv(cfg.INPUT_SIZE, 2)
pad_right = tf.math.floordiv(pad_size, 2) + tf.math.floordiv(cfg.INPUT_SIZE, 2)
if tf.math.mod(pad_size, 2) > 0:
pad_right += 1
# Pad By Concatenating Left/Right Edge Values
data = self.pad_edge(data, pad_left, 'LEFT')
data = self.pad_edge(data, pad_right, 'RIGHT')
# Pad Non Empty Frame Indices
non_empty_frames_idxs = self.pad_edge(non_empty_frames_idxs, pad_left, 'LEFT')
non_empty_frames_idxs = self.pad_edge(non_empty_frames_idxs, pad_right, 'RIGHT')
# Reshape to Mean Pool
data = tf.reshape(data, [cfg.INPUT_SIZE, -1, N_COLS, cfg.N_DIMS])
non_empty_frames_idxs = tf.reshape(non_empty_frames_idxs, [cfg.INPUT_SIZE, -1])
# Mean Pool
data = tf.experimental.numpy.nanmean(data, axis=1)
non_empty_frames_idxs = tf.experimental.numpy.nanmean(non_empty_frames_idxs, axis=1)
# Fill NaN Values With 0
data = tf.where(tf.math.is_nan(data), 0.0, data)
return data, non_empty_frames_idxs
preprocess_layer = PreprocessLayer()load_relevant_data_subset is to load a subset of relevant data from a parquet file . The data is then reshaped into a 3D numpy array.
# Source: https://www.kaggle.com/competitions/asl-signs/overview/evaluation
ROWS_PER_FRAME = 543 # number of landmarks per frame
def load_relevant_data_subset(pq_path):
data_columns = ['x', 'y', 'z']
data = pd.read_parquet(pq_path, columns=data_columns)
n_frames = int(len(data) / ROWS_PER_FRAME)
data = data.values.reshape(n_frames, ROWS_PER_FRAME, len(data_columns))
return data.astype(np.float32)This function loads a subset of the relevant data from the given file path by calling the load_relevant_data_subset function. Then it processes the data using TensorFlow by using the preprocess_layer function.
"""
face: 0:468
left_hand: 468:489
pose: 489:522
right_hand: 522:544
"""
def get_data(file_path):
# Load Raw Data
data = load_relevant_data_subset(file_path)
# Process Data Using Tensorflow
data = preprocess_layer(data)
return dataCreate Dataset
preprocess_data function generates and saves training and validation data for a sign language classification task. The function creates several numpy arrays to store the data, including X and y arrays to hold the input data and labels. For each row in the train DataFrame, the function calls a get_data function to load and preprocess the data from the file specified by the file_path column, and then stores the resulting data and labels in the appropriate arrays. The function also creates a 2D array called NON_EMPTY_FRAME_IDXS to store the indices of non-empty frames for each sample. After generating and storing all the data, the function splits it into training and validation sets.
# Get the full dataset
def preprocess_data():
# Create arrays to save data
X = np.zeros([N_SAMPLES, cfg.INPUT_SIZE, N_COLS, cfg.N_DIMS], dtype=np.float32)
y = np.zeros([N_SAMPLES], dtype=np.int32)
NON_EMPTY_FRAME_IDXS = np.full([N_SAMPLES, cfg.INPUT_SIZE], -1, dtype=np.float32)
# Fill X/y
for row_idx, (file_path, sign_ord) in enumerate(tqdm(train[['file_path', 'sign_ord']].values)):
# Log message every 5000 samples
if row_idx % 5000 == 0:
print(f'Generated {row_idx}/{N_SAMPLES}')
data, non_empty_frame_idxs = get_data(file_path)
X[row_idx] = data
y[row_idx] = sign_ord
NON_EMPTY_FRAME_IDXS[row_idx] = non_empty_frame_idxs
# Sanity check, data should not contain NaN values
if np.isnan(data).sum() > 0:
print(row_idx)
return data
# Save X/y
np.save('X.npy', X)
np.save('y.npy', y)
np.save('NON_EMPTY_FRAME_IDXS.npy', NON_EMPTY_FRAME_IDXS)
# Save Validation
splitter = GroupShuffleSplit(test_size=0.10, n_splits=2, random_state=cfg.SEED)
PARTICIPANT_IDS = train['participant_id'].values
train_idxs, val_idxs = next(splitter.split(X, y, groups=PARTICIPANT_IDS))
# Save Train
X_train = X[train_idxs]
NON_EMPTY_FRAME_IDXS_TRAIN = NON_EMPTY_FRAME_IDXS[train_idxs]
y_train = y[train_idxs]
np.save('X_train.npy', X_train)
np.save('y_train.npy', y_train)
np.save('NON_EMPTY_FRAME_IDXS_TRAIN.npy', NON_EMPTY_FRAME_IDXS_TRAIN)
# Save Validation
X_val = X[val_idxs]
NON_EMPTY_FRAME_IDXS_VAL = NON_EMPTY_FRAME_IDXS[val_idxs]
y_val = y[val_idxs]
np.save('X_val.npy', X_val)
np.save('y_val.npy', y_val)
np.save('NON_EMPTY_FRAME_IDXS_VAL.npy', NON_EMPTY_FRAME_IDXS_VAL)
# Split Statistics
print(f'Patient ID Intersection Train/Val: {set(PARTICIPANT_IDS[train_idxs]).intersection(PARTICIPANT_IDS[val_idxs])}')
print(f'X_train shape: {X_train.shape}, X_val shape: {X_val.shape}')
print(f'y_train shape: {y_train.shape}, y_val shape: {y_val.shape}')If cfg.PREPROCESS_DATA is True, preprocess the data by calling a function preprocess_data().
# Preprocess All Data From Scratch
if cfg.PREPROCESS_DATA:
preprocess_data()
ROOT_DIR = '.'
else:
ROOT_DIR = '/content/drive/MyDrive/23_Google/data/gislr-dataset-public'
# Load Data
if cfg.USE_VAL:
# Load Train
X_train = np.load(f'{ROOT_DIR}/X_train.npy')
y_train = np.load(f'{ROOT_DIR}/y_train.npy')
NON_EMPTY_FRAME_IDXS_TRAIN = np.load(f'{ROOT_DIR}/NON_EMPTY_FRAME_IDXS_TRAIN.npy')
# Load Val
X_val = np.load(f'{ROOT_DIR}/X_val.npy')
y_val = np.load(f'{ROOT_DIR}/y_val.npy')
NON_EMPTY_FRAME_IDXS_VAL = np.load(f'{ROOT_DIR}/NON_EMPTY_FRAME_IDXS_VAL.npy')
# Define validation Data
validation_data = ({ 'frames': X_val, 'non_empty_frame_idxs': NON_EMPTY_FRAME_IDXS_VAL }, y_val)
else:
X_train = np.load(f'{ROOT_DIR}/X.npy')
y_train = np.load(f'{ROOT_DIR}/y.npy')
NON_EMPTY_FRAME_IDXS_TRAIN = np.load(f'{ROOT_DIR}/NON_EMPTY_FRAME_IDXS.npy')
validation_data = None
# Train
print_shape_dtype([X_train, y_train, NON_EMPTY_FRAME_IDXS_TRAIN], ['X_train', 'y_train', 'NON_EMPTY_FRAME_IDXS_TRAIN'])
# Val
if cfg.USE_VAL:
print_shape_dtype([X_val, y_val, NON_EMPTY_FRAME_IDXS_VAL], ['X_val', 'y_val', 'NON_EMPTY_FRAME_IDXS_VAL'])
# Sanity Check
print(f'# NaN Values X_train: {np.isnan(X_train).sum()}')
'''
X_train shape: (94477, 64, 66, 3), dtype: float32
y_train shape: (94477,), dtype: int32
NON_EMPTY_FRAME_IDXS_TRAIN shape: (94477, 64), dtype: float32
# NaN Values X_train: 0
'''The count of each class in the y_train data is computed and displayed. The iloc method is used to select the first and last five classes from the DataFrame and display them.
# Class Count
display(pd.Series(y_train).value_counts().to_frame('Class Count').iloc[[0,1,2,3,4, -5,-4,-3,-2,-1]])
"""
Class Count
135 415
136 414
194 411
60 410
148 408
56 312
170 312
21 310
231 307
249 299
"""Number Of Frames
A waterfall plot is created to show the percentage of samples in a dataset that have at least a certain number of non-empty frames.
# Vast majority of samples fits has less than 32 non empty frames
N_EMPTY_FRAMES = (NON_EMPTY_FRAME_IDXS_TRAIN != -1).sum(axis=1)
N_EMPTY_FRAMES_WATERFALL = []
for n in tqdm(range(1,cfg.INPUT_SIZE+1)):
N_EMPTY_FRAMES_WATERFALL.append(sum(N_EMPTY_FRAMES >= n) / len(NON_EMPTY_FRAME_IDXS_TRAIN) * 100)
plt.figure(figsize=(18,10))
plt.title('Waterfall Plot For Number Of Non Empty Frames')
pd.Series(N_EMPTY_FRAMES_WATERFALL).plot(kind='bar')
plt.grid(axis='y')
plt.xticks(np.arange(cfg.INPUT_SIZE), np.arange(1, cfg.INPUT_SIZE+1))
plt.xlabel('Number of Non Empty Frames', size=16)
plt.yticks(np.arange(0, 100+10, 10))
plt.ylim(0, 100)
plt.ylabel('Percentage of Samples With At Least N Non Empty Frames', size=16)
plt.show()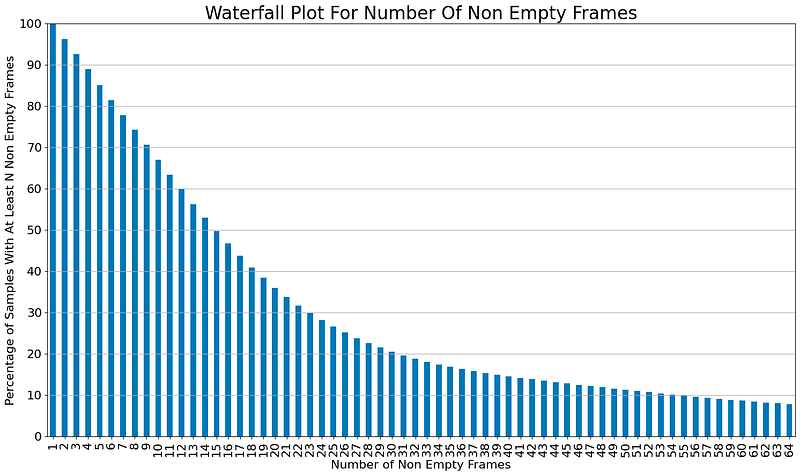
Percentage of Frames Filled
In order to know the percentage of frames that are filled in the training dataset, we need to calculate the maximum fill percentage of each landmark by counting the number of non-empty frames (frames with data) for each landmark and dividing it by the total number of frames in the training dataset. Each landmark has 31.46% of its frames filled with data.
# Percentage of frames filled, this is the maximum fill percentage of each landmark
P_DATA_FILLED = (NON_EMPTY_FRAME_IDXS_TRAIN != -1).sum() / NON_EMPTY_FRAME_IDXS_TRAIN.size * 100
print(f'P_DATA_FILLED: {P_DATA_FILLED:.2f}%')
'''
P_DATA_FILLED: 31.46%
'''Mean and standard deviations
We are going to calculate means and standard deviations for 3 types of landmarks: Lips, Hands, and Pose. Calculating the mean and standard deviation is an important step in normalizing the data. For getting the percentage of left lips measurements in the training data, we count the number of non-zero values in the left lips landmarks for all frames and divide it by the total number of elements in the left lips landmarks for all frames.
# Percentage of Lips Measurements
P_LEFT_LIPS_MEASUREMENTS = (X_train[:,:,LIPS_IDXS] != 0).sum() / X_train[:,:,LIPS_IDXS].size / P_DATA_FILLED * 1e4
print(f'P_LEFT_LIPS_MEASUREMENTS: {P_LEFT_LIPS_MEASUREMENTS:.2f}%')
'''
P_LEFT_LIPS_MEASUREMENTS: 99.40%
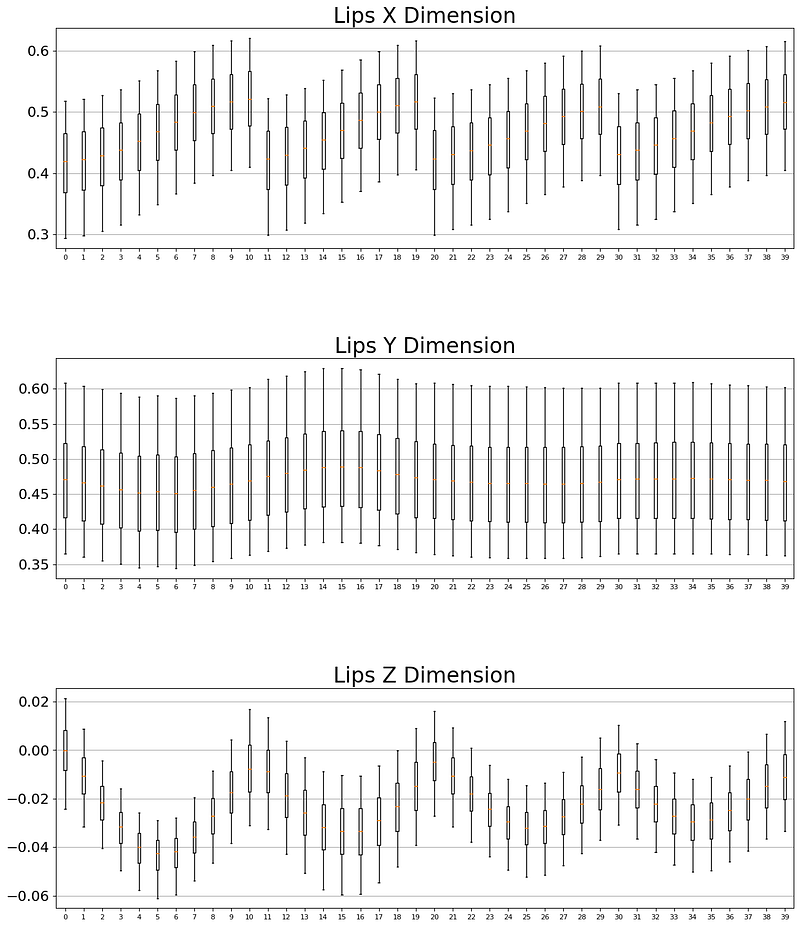
'''We need to calculate the mean and standard deviation of the lip landmark positions in the training dataset for normalization. We can create three subplots, one for each dimension of the lip landmarks (x, y, and z), and generate a box plot of the non-zero values of each landmark position for each dimension. The box plots are used to visualize the distribution of the data and to identify outliers. Finally, the function returns the mean and standard deviation of the non-zero lip landmark positions.
def get_lips_mean_std():
# LIPS
LIPS_MEAN_X = np.zeros([LIPS_IDXS.size], dtype=np.float32)
LIPS_MEAN_Y = np.zeros([LIPS_IDXS.size], dtype=np.float32)
LIPS_STD_X = np.zeros([LIPS_IDXS.size], dtype=np.float32)
LIPS_STD_Y = np.zeros([LIPS_IDXS.size], dtype=np.float32)
fig, axes = plt.subplots(3, 1, figsize=(15, cfg.N_DIMS*6))
for col, ll in enumerate(tqdm( np.transpose(X_train[:,:,LIPS_IDXS], [2,3,0,1]).reshape([LIPS_IDXS.size, cfg.N_DIMS, -1]) )):
for dim, l in enumerate(ll):
v = l[np.nonzero(l)]
if dim == 0: # X
LIPS_MEAN_X[col] = v.mean()
LIPS_STD_X[col] = v.std()
if dim == 1: # Y
LIPS_MEAN_Y[col] = v.mean()
LIPS_STD_Y[col] = v.std()
axes[dim].boxplot(v, notch=False, showfliers=False, positions=[col], whis=[5,95])
for ax, dim_name in zip(axes, cfg.DIM_NAMES):
ax.set_title(f'Lips {dim_name.upper()} Dimension', size=24)
ax.tick_params(axis='x', labelsize=8)
ax.grid(axis='y')
plt.subplots_adjust(hspace=0.50)
plt.show()
LIPS_MEAN = np.array([LIPS_MEAN_X, LIPS_MEAN_Y]).T
LIPS_STD = np.array([LIPS_STD_X, LIPS_STD_Y]).T
return LIPS_MEAN, LIPS_STD
LIPS_MEAN, LIPS_STD = get_lips_mean_std()In this article, I only show the case of the lip because the process is the same for the other two landmarks.
Custom Sampler
The custom sampler function is used to get a batch containing all samples for all classes (or a specific class) N times. The function randomly selects data samples in batches of size cfg.BATCH_ALL_SIGNS_N and creates a batch that contains all samples for all classes. Within a loop, the function creates a dictionary called CLASS2IDXS that stores the indices of samples that belong to each class.
# Custom sampler to get a batch containing N times all signs
def get_train_batch_all_signs(X, y, NON_EMPTY_FRAME_IDXS, n=cfg.BATCH_ALL_SIGNS_N):
# Arrays to store batch in
X_batch = np.zeros([cfg.NUM_CLASSES*n, cfg.INPUT_SIZE, N_COLS, cfg.N_DIMS], dtype=np.float32)
y_batch = np.arange(0, cfg.NUM_CLASSES, step=1/n, dtype=np.float32).astype(np.int64)
non_empty_frame_idxs_batch = np.zeros([cfg.NUM_CLASSES*n, cfg.INPUT_SIZE], dtype=np.float32)
# Dictionary mapping ordinally encoded sign to corresponding sample indices
CLASS2IDXS = {}
for i in range(cfg.NUM_CLASSES):
CLASS2IDXS[i] = np.argwhere(y == i).squeeze().astype(np.int32)
while True:
# Fill batch arrays
for i in range(cfg.NUM_CLASSES):
idxs = np.random.choice(CLASS2IDXS[i], n)
X_batch[i*n:(i+1)*n] = X[idxs]
non_empty_frame_idxs_batch[i*n:(i+1)*n] = NON_EMPTY_FRAME_IDXS[idxs]
yield { 'frames': X_batch, 'non_empty_frame_idxs': non_empty_frame_idxs_batch }, y_batchLet’s check with a dummy dataset.
dummy_dataset = get_train_batch_all_signs(X_train, y_train, NON_EMPTY_FRAME_IDXS_TRAIN)
X_batch, y_batch = next(dummy_dataset)
for k, v in X_batch.items():
print(f'{k} shape: {v.shape}, dtype: {v.dtype}')
# Batch shape/dtype
print(f'y_batch shape: {y_batch.shape}, dtype: {y_batch.dtype}')
# Verify each batch contains each sign exactly N times
display(pd.Series(y_batch).value_counts().to_frame('Counts'))
"""
frames shape: (1000, 64, 66, 3), dtype: float32
non_empty_frame_idxs shape: (1000, 64), dtype: float32
y_batch shape: (1000,), dtype: int64
"""In this article, we process the dataset for training. In the next article, we will deal with the transformer model and how to train it.
Thank you for reading!
